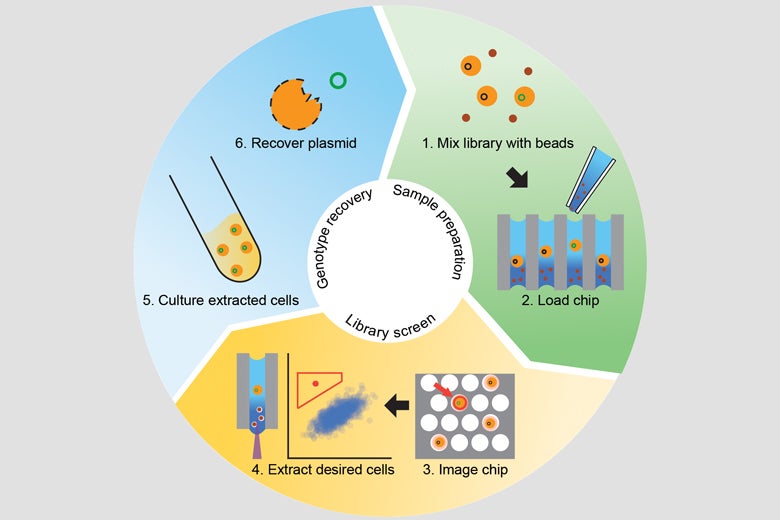
An overview of the directed evolution process using the new Stanford technique: preparing protein libraries, screening them, extracting desired cells, and then inferring their DNA sequence. (Click image to enlarge.) (Image credit: Cochran Lab)
All living things require proteins, members of a vast family of molecules that nature “makes to order” according to the blueprints in DNA.
Through the natural process of evolution, DNA mutations generate new or more effective proteins. Humans have found so many alternative uses for these molecules – as foods, industrial enzymes, anti-cancer drugs – that scientists are eager to better understand how to engineer protein variants designed for specific uses.
Now Stanford engineers have invented a technique to dramatically accelerate protein evolution for this purpose. This technology, described in Nature Chemical Biology, allows researchers to test millions of variants of a given protein, choose the best for some task and determine the DNA sequence that creates this variant.
“Evolution, the survival of the fittest, takes place over a span of thousands of years, but we can now direct proteins to evolve in hours or days,” said Jennifer Cochran, an associate professor of bioengineering who co-authored the paper with Thomas Baer, executive director of the Stanford Photonics Research Center.
“This is a practical, versatile system with broad applications that researchers will find easy to use,” Baer said.
By combining Cochran’s protein engineering know-how with Baer’s expertise in laser-based instrumentation, the team created a tool that can test millions of protein variants in a matter of hours.
“The demonstrations are impressive and I look forward to seeing this technology more widely adopted,” said Frances Arnold, a professor of chemical engineering at Caltech who was not affiliated with the study.
Making a million mutants
The researchers call their tool µSCALE, or Single Cell Analysis and Laser Extraction.
The “µ” stands for the microcapillary glass slide that holds the protein samples. The slide is roughly the size and thickness of a penny, yet in that space a million capillary tubes are arrayed like straws, open on the top and bottom.
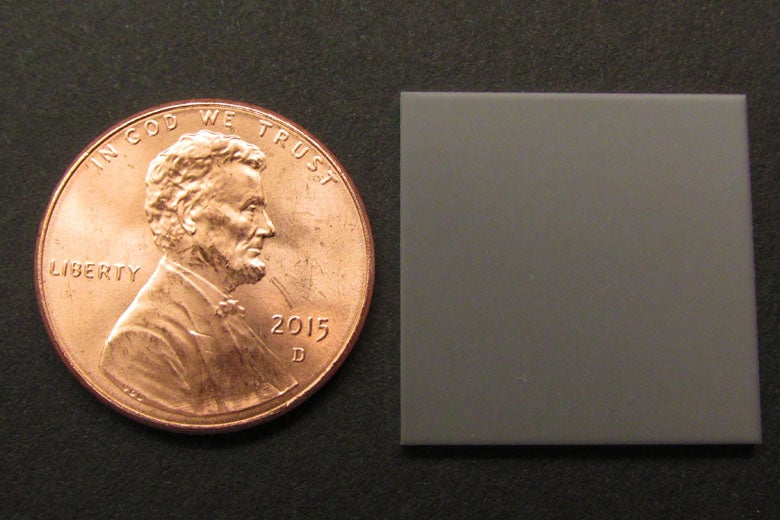
The microcapillary glass slide, roughly the size and thickness of a penny, holds the protein samples. (Image credit: Courtesy Cochran Lab)
The power of µSCALE is how it enables researchers to build upon current biochemical techniques to run a million protein experiments simultaneously, then extract and further analyze the most promising results.
The researchers first employ a process termed “mutagenesis” to create random variations in a specific gene. These mutations are inserted into batches of yeast or bacterial cells, which express the altered gene and produce millions of random protein variants.
A µSCALE user mixes millions of tiny opaque glass beads into a sample containing millions of yeast or bacteria and spreads the mixture on a microcapillary slide. Tiny amounts of fluid trickle into each tube, carrying individual cells. Surface tension traps the liquid and the cell in each capillary.
The slide bearing these million yeast or bacteria, and the protein variants they produce, is inserted into the µSCALE device. A software-controlled microscope peers into each capillary and takes images of the biochemical reaction occurring therein.
Once a µSCALE user identifies a capillary of interest, the researcher can direct the laser to extract the contents of that tube without disrupting its neighbors, using an ingenious method devised by Baer.
“The beads are what enable extraction,” Baer said. “The laser supplies energy to move the beads, which breaks the surface tension and releases the sample from the capillary.”
Thus µSCALE empties the contents of a single capillary onto a collector plate, where the DNA of the isolated cell can be sequenced and the gene variant responsible for the protein of interest can be identified.
“One of the unique features of µSCALE is that it allows researchers to rapidly isolate a single desired cell from hundreds of thousands of other cells,” said Bob Chen, a doctoral student in Cochran’s lab who wrote the software to examine and detect signs of interesting protein activity within the test tubes.
Promising variants can be collected and reprocessed through µSCALE to further evolve and optimize the protein.
“This is an exciting new tool to answer important questions about proteins,” Cochran said, likening µSCALE to the way that high-throughput tools for gene analysis have allowed researchers to unlock key features of biology underlying human disease.
Genesis and proofs
The project began five years ago when Baer and collaborator Ivan Dimov developed the first instrument. They showed how to identify cell types in a microcapillary array and extract a single capillary’s contents using glass beads and a focused laser.
About three years ago, Cochran and Baer joined forces to develop µSCALE for protein engineering, and the team devised three experiments to showcase µSCALE’s utility and flexibility.
In one experiment, researchers sifted through a protein library produced in yeast cells to select antibodies that bound most tightly to a cancer target. Antibodies with a high target-binding affinity are known to be effective against cancer.
In a second example, they engineered a bright orange fluorescent protein biosensor. Using µSCALE, they did this almost 10 times faster than previous methods. Such biosensors are often used as tags in a wide variety of biology experiments.
A third experiment, carried out with Stanford biochemistry Professor Daniel Herschlag, used µSCALE to improve upon a model enzyme.
“This system will allow us to explore the evolutionary and functional relationships between enzymes, guiding the engineering of new enzymes that can carry out novel beneficial reactions,” Herschlag said.
The Stanford team included graduate students Sungwon Lim and Arvind Kannan, postdoctoral scholar Spencer Alford and researcher Fanny Sunden.
Support for this work included a Wallace H. Coulter Translational Partnership Award, which made this pioneering interdisciplinary research possible.
Media Contacts
Tom Abate, Stanford Engineering: (650) 736-2245, tabate@stanford.edu
Bjorn Carey, Stanford News Service: (650) 725-1944, bccarey@stanford.edu