|
December 12, 2012
Stanford experiment finds ulcer bug's weak point
SLAC's high-power X-rays have revealed a potential drug target in H. pylori, the ulcer-causing bacteria that infect half the world's population. 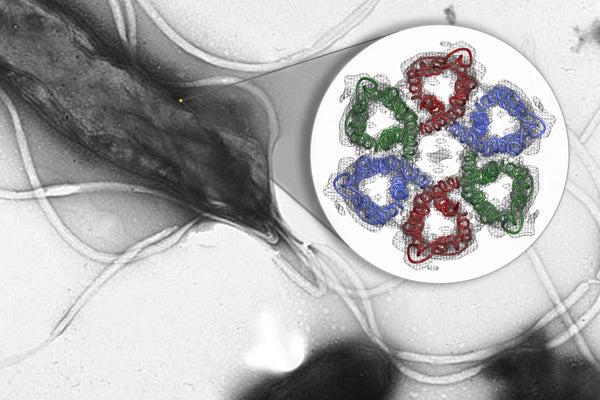
This diagram shows the first-ever glimpse of the six-part ring of urea channels embedded in the membrane of a type of bacteria called Helicobacter pylori. (Image: Hartmut Luecke / UC-Irvine)
In 1982, Australian scientists extracted bacteria from a person's stomach, grew them in a petri dish and identified them as the cause of ulcers and gastritis. Three decades later, scientists have now used powerful X-rays at the SLAC National Accelerator Laboratory at Stanford to reveal a potential way to attack the common stomach bacteria.
At least half the world's population carries the bacteria, Helicobacter pylori, and hundreds of millions suffer health problems that ultimately increase the odds of developing stomach cancer. Current treatments require a complicated regimen of stomach-acid inhibitors and antibiotics, the latter of which have the side effect of indiscriminately knocking out beneficial bacteria.
H. pylori is a robust bacterium, able to thrive in an environment that's as caustic as car battery acid. Crucial to H. pylori's survival are tiny protein channels within its cell membrane. Urea from the surrounding gastric juices passes through these channels and into the bacterium, which converts the urea into ammonia that protects it from the acid.
Blocking the channels would disable this protective system, leading to a new treatment for people with the infection.
Using X-rays from SLAC's Stanford Synchrotron Radiation Lightsource (SSRL), "we have deciphered the three-dimensional molecular structure of a very promising drug target," said Hartmut "Hudel" Luecke, a researcher at the University of California-Irvine and principal investigator on the paper, published online Dec. 9 in Nature.
Solving the structure of the protein to find the specific area to target wasn't easy. It is notoriously difficult to crystallize membrane proteins, which is a prerequisite step for using protein crystallography, the primary technique for visualizing protein structures. This technique bounces X-rays off of the electrons in the crystallized protein, which generates the experimental data used to build a 3-D map of the protein's atoms.
"We collected over 100 separate data sets and tried numerous structural determination techniques," said Mike Soltis, head of SSRL's structural molecular biology group, who worked with Luecke and his team to create the 3-D map of the atomic structure.
"This is the hardest structure I've ever deciphered, and I've been doing this since 1984," Luecke said. "You have to try all kinds of tricks, and these crystals fought us every step of the way. But now that we have the structure, we've reached the exciting part – the prospect of creating specific, safe and effective ways to target this pathogen and wipe it out."
This work was supported by the National Institutes of Health, the National Cancer Institute, the UC-Irvine Center for Biomembrane Systems and the U.S. Veterans Administration.
The research paper is available online from Nature.
-30-
|